
 |
Manual |
All DMD jobs should be targeted to the Alamo cluster, where there is no run time limit.
Cluster usage is a work in progress and errors in cluster jobs are currently not directly reported.
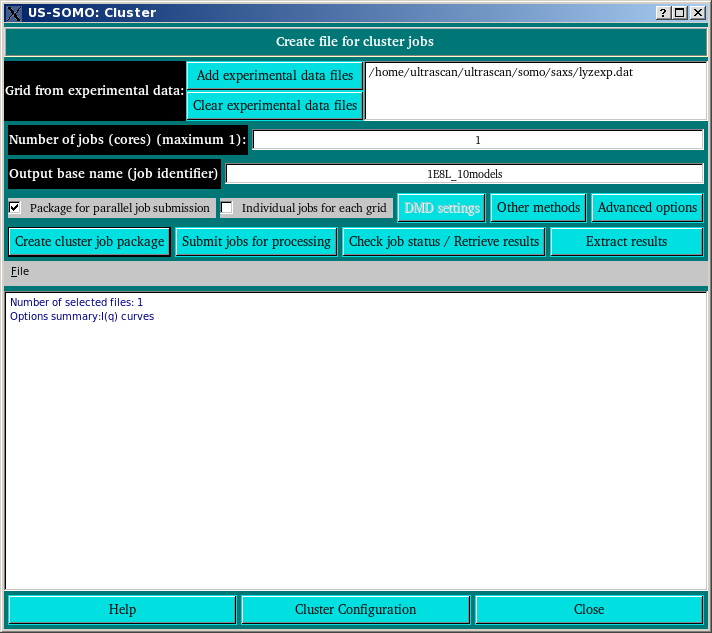
This module allows to pack files into structured .tar archives for submission to local/remote (super)computing cluster(s).
In the Grid from experimental data: section, by pressing the Add experimental data files button you can pick from your filesystem an experimentallly-derived SAXS I(q) vs q curve that will be used to set the q-range and stepsize over which the computations will be carried on. The Clear experimental data files button will allow the removal of unwanted files from the list.
The Number of jobs (maximum n): field is set to 1 on opening the module and then adjusted to the number of independent structures in the file when the Package for parallel job submission checkbox is selected. It can be subsequently manually edited to change the number of jobs up to the maximum value shown if desired. Changing the number of Grids from experimental data, Advanced Options, and DMD Settings can effect the maximum number of jobs possible. Depending on the limits and characteristics of the target cluster, you may wish to use the maximum possible value or something less. Each of the jobs will be processed in parallel, up to the limits of the target cluster. Using more processors will result in less time taken to run the job, but may take more time waiting before the resources are available to run the job.
Write the output filename in the Output base name (job identifier) field. The system will check for the existance of identically-named jobs and will refuse to proceed until an unique name is inserted.
For DMD jobs, the DMD settings button will open up the DMD set-up panel.
The Other methods button will open up a pop-up panel where currently (April 2014) only the BEST hydrodynamic computations method can be selected (default: unselected).
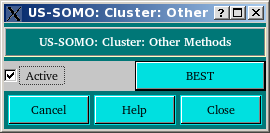
Pressing BEST will open the BEST settings page. Pressing Close will return to the cluster module main panel.
The Advanced options button will open up another pop-up window where multiple I(q) vs. q methods and other features can be selected (see here).
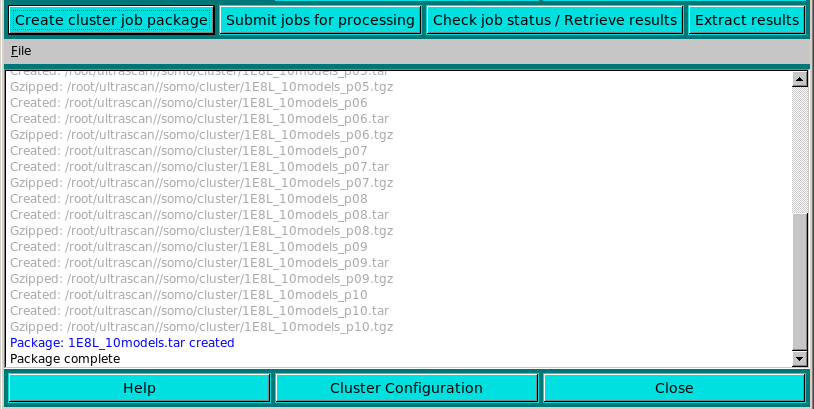
By pressing the Create cluster job package button, the .tar archive is prepared, with the intermediate operations reported in light grey in the progress window (see above).
The job submission to a cluster is then done by pressing the Submit jobs for processing button, which will open up the submission module.
Once one or multiple jobs have been submitted, their status can be checked, and results retrived by pressing the Check job status/Retrieve results button.
Finally, the results can be extracted by pressing the Extract results button, which will open up the extraction module).
Clusters details and configuration can be accessed by pressing the Cluster Configuration button.
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on April 24, 2014